In this post I will use fairly new R package sparklyr which allows you to connect from Spark with R to use Spark’s machine learning library. All with a complete dplyr backend. To connect to a local instance of Spark, you first need to spark_install and then connect to that instance with spark_connect where the master argument should be “local”
I will use the flights data from nycflights13 package to try to predict flights that will be delayed. As I import the data, I also create a new weekday variable
library(sparklyr)
library(dplyr)
sc <- spark_connect(master = "local", version="2.1.0")flights_data <- nycflights13::flights %>%
mutate(weekday=weekdays(as.Date(paste0(year,"-",month,"-",day))))To upload this data frame into my local instance, use copy_to()
flights_sdf <- copy_to(dest = sc, df = flights_data, name = "name", overwrite=T)Then, I create a new variable i) Delay is 1 if the plane was delayed, ii) Car is 1 if the flights is flown by one of the four selected carreirs, iii) Night that is 1 if the the departure was after 6 pm or earlier than 6am and iv) Weekendif the departure was on a Sunday.
flights_sdf = flights_sdf %>%
mutate(Delay = if_else(dep_delay>0,1,0)) %>% # (i)
mutate(Car = if_else(carrier %in% c("DL","US","DH","UA"),1,0)) %>% # (ii)
mutate(Night = if_else(hour>=18 | hour<=6,1,0)) %>% # (iii)
mutate(Weekend = if_else(weekday=="Sunday",1,0)) # (iv)I create a new data frame logit_sdf with the constructed variables, and exluce all missing values
logit_sdf = flights_sdf %>%
mutate(DepHour = hour) %>%
select(Delay, Car, Night, Weekend, DepHour) %>%
na.omit()## * Dropped 8255 rows with 'na.omit' (336776 => 328521)I divide the dataset into two parts, one for training and one for testing, and then fit the binary logistic regression model with the Spark Machine Learning function ml_logistic_regression
partitions <- sdf_partition(logit_sdf, training=0.7, test=0.3, seed=6789)
trainset <- partitions$training
testset <- partitions$test
ml_model <- trainset %>% ml_logistic_regression(formula = Delay ~ DepHour + Car + Night + Weekend)
summary(ml_model)## Call: ml_logistic_regression.tbl_spark(., formula = Delay ~ DepHour + Car + Night + Weekend)
##
## Coefficients:
## (Intercept) DepHour Car Night Weekend
## -1.88682995 0.11464268 -0.06620449 -0.19125631 -0.08851651Prediction is done with sdf_predict. Also a matrix with TP = True positive, FP = False positive, TN = True negative, FN = False negative. For now, I consider the cut-off value for when we would classify a predicted probability as a delay to be 0.5. The model falsely predict roughly 29 thousand flights as delayed when they were in fact not.
predvals <- sdf_predict(x = testset,model = ml_model)
score_test <- predvals %>%
mutate(Event = if_else(probability_1_0 >= 0.5,1,0)) %>%
mutate(TP = if_else(Event==1 & Delay==1, 1, 0)) %>%
mutate(FP = if_else(Event==1 & Delay==0, 1, 0)) %>%
mutate(FN = if_else(Event==0 & Delay==1, 1, 0)) %>%
mutate(TN = if_else(Event==0 & Delay==0, 1, 0))%>%
summarise(nTP = sum(TP, na.rm=T),
nFP = sum(FP, na.rm=T),
nFN = sum(FN, na.rm=T),
nTN = sum(TN, na.rm=T)) %>%
select(nTP,nFP,nFN,nTN)
mat <- t(matrix(score_test %>% collect(),ncol=2,byrow=T,dimnames=list(
c("TRUE", "FALSE"),
c("Predicted: TRUE", "Predicted: FALSE")
)))
mat## TRUE FALSE
## Predicted: TRUE 9640 28893
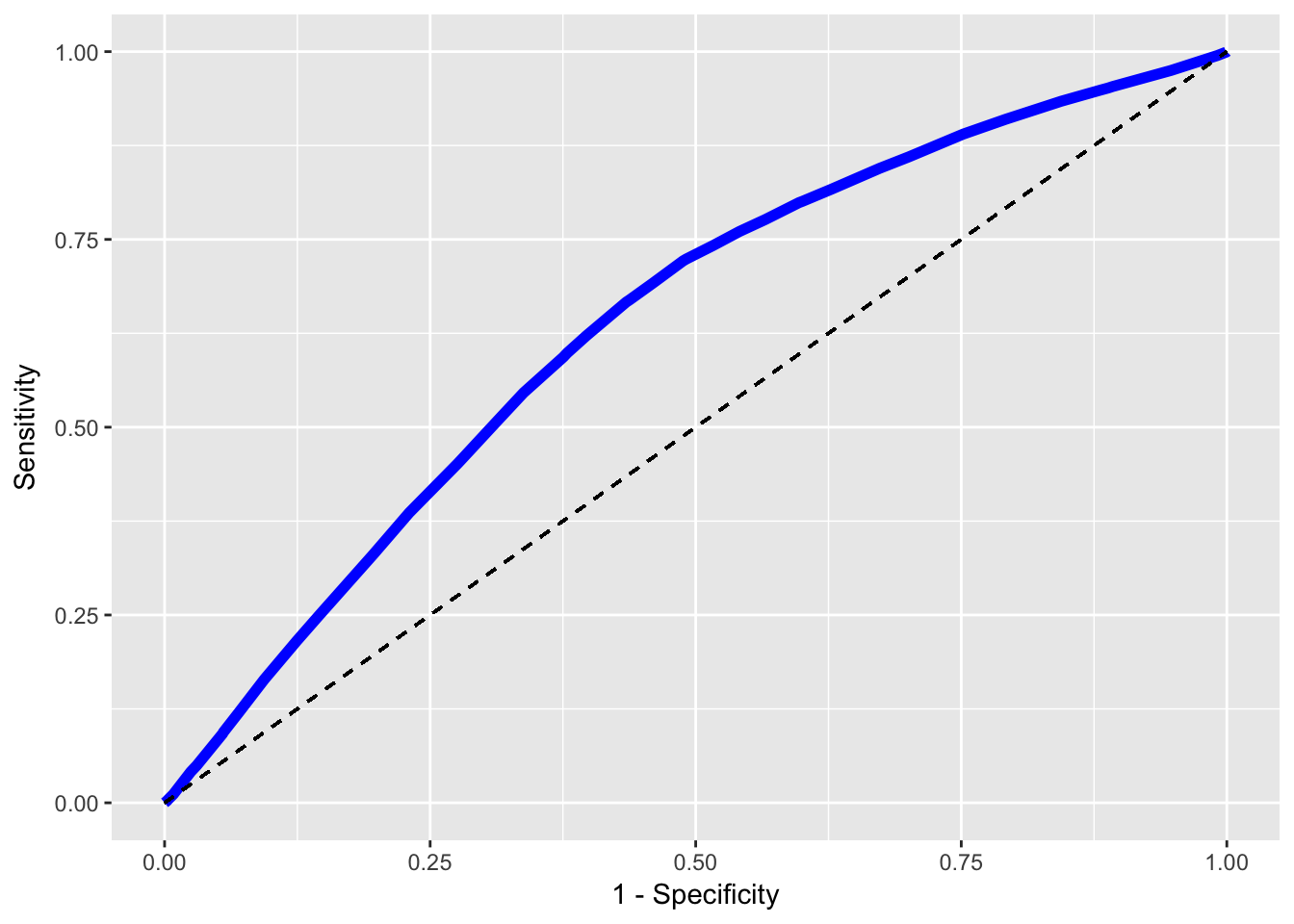
## Predicted: FALSE 8734 51136Instead of using a specific cut-off value as above, it is better to look at a ROC-curve. Here I use the previous code to construct a function calc_roc that will output the sensitivity and specificity of the model given a specified cut-off value. I will then loop through a sequence of different cut-off values and see how sensitivity and specificity changes for different cut-off values. A much more rigorous method of evaluating the model.
calc_roc <- function(x, k=0.5){
x <- x %>%
mutate(Event = if_else(probability_1_0 >= k, 1, 0)) %>%
mutate(TP = if_else(Event == 1 & Delay==1, 1, 0)) %>%
mutate(TN = if_else(Event == 0 & Delay==0, 1, 0)) %>%
mutate(P = if_else(Delay == 1, 1, 0 )) %>%
mutate(N = if_else(Delay == 0, 1, 0)) %>%
summarise(n1 = sum(TP, na.rm = T),
n2 = sum(P, na.rm = T),
n3 = sum(TN, na.rm = T),
n4 = sum(N, na.rm = T)) %>%
summarise(val1 = n1/n2,
val2 = n3/n4) %>%
select(Sensitivity = val1, Specificity = val2)
return(x)
}
pred <- predvals %>% collect
perf<-vector()
for(i in seq(0,1,by=0.01)){
perf <- rbind(perf, do.call(calc_roc, list(pred,i)))
}
colnames(perf) <- c("Sensitivity","Specificity")The result can then be plotted using ggplot2.
library(ggplot2)
ggplot(perf, aes(x=1-Specificity, y=Sensitivity))+
geom_line(col="blue",lwd=2)+
geom_segment(aes(x=0,xend=1,y=0,yend=1),linetype=2)